---
title: "Introduction to metalite.ae"
output: rmarkdown::html_vignette
vignette: >
%\VignetteIndexEntry{Introduction to metalite.ae}
%\VignetteEngine{knitr::rmarkdown}
%\VignetteEncoding{UTF-8}
resource_files:
- fig/*.png
- rtf/*.rtf
- pdf/*.pdf
---
```{r, include=FALSE}
knitr::opts_chunk$set(
comment = "#>",
collapse = TRUE,
out.width = "100%",
dpi = 150,
eval = TRUE
)
```
```{r, include=FALSE}
library(metalite.ae)
```
## Overview
metalite.ae is an R package designed for the analysis of adverse events (AE)
in clinical trials.
It operates on ADaM datasets and adheres to the metalite structure.
The package encompasses the following components:
AE summary.
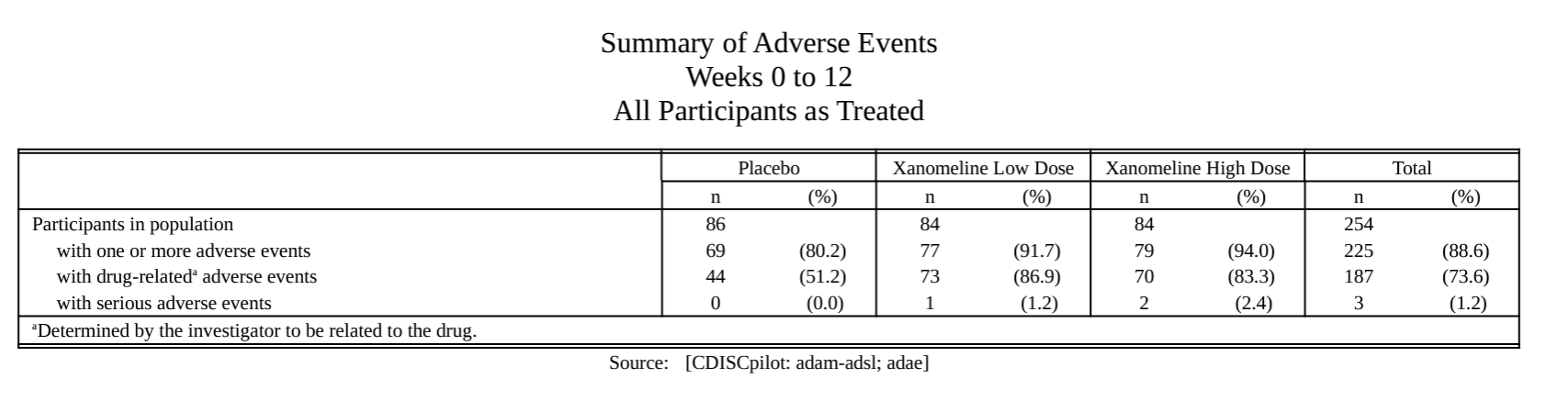
Specific AE analysis.
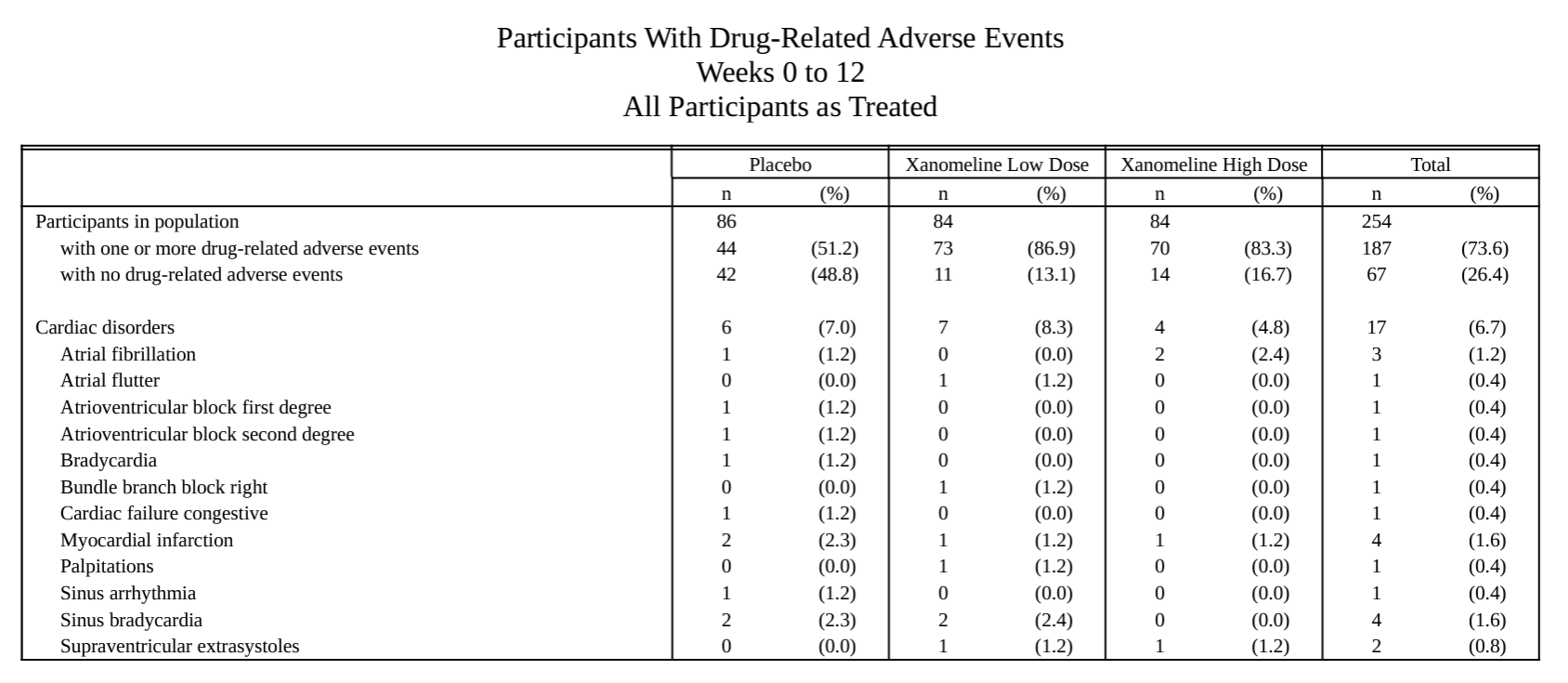
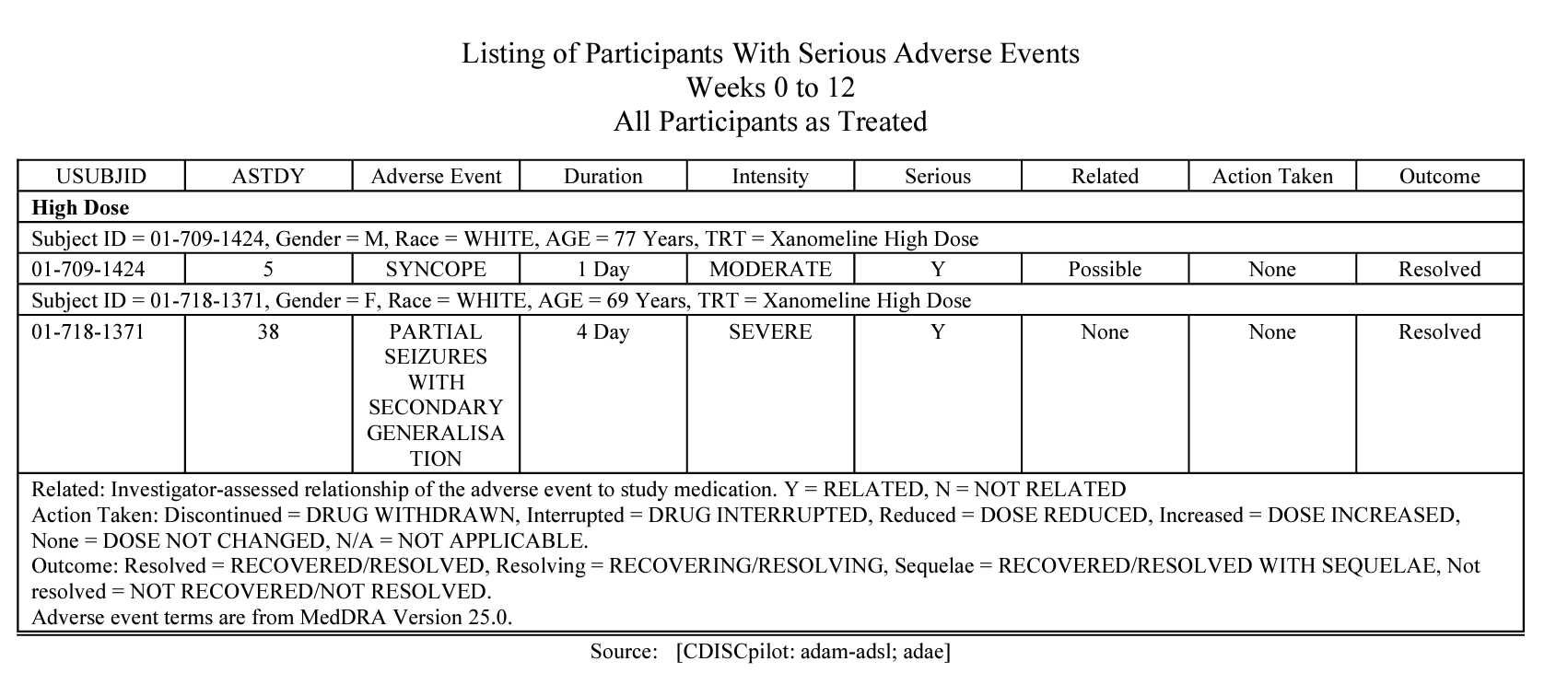
AE listing.
The R package streamlines the process of generating production-ready tables,
listings, and figures as outlined in the
[AE summary chapter](https://r4csr.org/tlf-ae-summary.html) and the
[specific AE chapter](https://r4csr.org/tlf-ae-specific.html) of the
_R for Clinical Study Reports and Submission_ book.
It ensures complete traceability throughout the development lifecycle,
leveraging the metalite data structure.
This R package offers a comprehensive software development lifecycle (SDLC)
solution, encompassing activities such as definition, development,
validation, and finalization of the analysis.
## Highlighted features
- Avoid duplicated input by using metadata structure.
- For example, define analysis population once to use
in all adverse events analysis.
- Consistent input and output in standard functions.
- Streamlines mock table generation.
## Workflow
The overall workflow includes the following steps:
1. Define metadata information using metalite R package.
1. Prepare outdata using `prepare_*()` functions.
1. Extend outdata using `extend_*()` functions (optional).
1. Format outdata using `format_*()` functions.
1. Create TLFs using `tlf_*()` functions.
For instance, we can illustrate the creation of a straightforward
AE summary table as shown below.
```{r, eval = FALSE}
meta_ae_example() |> # Example AE data created using metalite
prepare_ae_summary(
population = "apat", # Select population by keywords
observation = "wk12", # Select observation by keywords
parameter = "any;rel;ser" # Select AE terms by keywords
) |>
format_ae_summary() |>
tlf_ae_summary(
source = "Source: [CDISCpilot: adam-adsl; adae]", # Define data source
analysis = "ae_specific", # Provide analysis type defined in meta$analysis
path_outtable = "ae0summary.rtf" # Define output
)
```
```{r, out.width = "100%", out.height = "400px", echo = FALSE, fig.align = "center"}
knitr::include_graphics("pdf/ae0summary1.pdf")
```
Additional examples and tutorials can be found on the
[package website](https://merck.github.io/metalite.ae/articles/),
offering further guidance and illustrations.
## Input
To implement the workflow in metalite.ae, it is necessary to establish
a metadata structure using the metalite R package.
For detailed instructions, please consult the
[metalite tutorial](https://merck.github.io/metalite/articles/metalite.html)
and refer to the source code of the function
[`meta_ae_example()`](https://github.com/Merck/metalite.ae/blob/main/R/meta_ae_example.R).



